import torch
import numpy as np
import matplotlib.pyplot as plt
from scipy.stats import qmc
from tqdm.auto import tqdm
import seaborn as sns
class DiffusionProblem:
"""Defines the diffusion problem parameters and exact solution"""
def __init__(self, device='cuda' if torch.cuda.is_available() else 'cpu'):
self.device = device
print(f"Using device: {device}")
def exact_solution(self, x, t):
"""Compute exact solution u(x,t) = sin(πx)exp(-t)"""
return torch.sin(np.pi * x) * torch.exp(-t)
def source_term(self, x, t):
"""Compute source term of the PDE"""
return torch.exp(-t) * (-torch.sin(np.pi * x) + np.pi**2 * torch.sin(np.pi * x))
class PINN(torch.nn.Module):
"""Physics-Informed Neural Network for solving diffusion equation"""
def __init__(self, problem, hidden_layers=4, neurons=50):
super().__init__()
self.problem = problem
layers = []
layers.append(torch.nn.Linear(2, neurons))
layers.append(torch.nn.Tanh())
for _ in range(hidden_layers):
layers.append(torch.nn.Linear(neurons, neurons))
layers.append(torch.nn.Tanh())
layers.append(torch.nn.Linear(neurons, 1))
self.network = torch.nn.Sequential(*layers)
self.to(problem.device)
def forward(self, x, t):
return self.network(torch.cat([x, t], dim=1))
def compute_pde_residual(self, x, t):
"""Compute PDE residual using automatic differentiation"""
x.requires_grad_(True)
t.requires_grad_(True)
u = self.forward(x, t)
u_t = torch.autograd.grad(u.sum(), t, create_graph=True)[0]
u_x = torch.autograd.grad(u.sum(), x, create_graph=True)[0]
u_xx = torch.autograd.grad(u_x.sum(), x, create_graph=True)[0]
return u_t - u_xx - self.problem.source_term(x, t)
class PointGenerator:
"""Generates training points using various sampling methods"""
def __init__(self, problem):
self.problem = problem
def generate_points(self, n_points, method='grid'):
if method == 'grid':
return self._grid_points(n_points)
elif method == 'random':
return self._random_points(n_points)
elif method in ['lhs', 'halton', 'sobol', 'hammersley']:
return self._qmc_points(n_points, method)
else:
raise ValueError(f"Unknown sampling method: {method}")
def _grid_points(self, n_points):
n_per_dim = int(np.sqrt(n_points))
x = np.linspace(-1, 1, n_per_dim)
t = np.linspace(0, 1, n_per_dim)
X, T = np.meshgrid(x, t)
return (torch.tensor(X.flatten()[:, None], dtype=torch.float32, device=self.problem.device),
torch.tensor(T.flatten()[:, None], dtype=torch.float32, device=self.problem.device))
def _random_points(self, n_points):
x = np.random.uniform(-1, 1, (n_points, 1))
t = np.random.uniform(0, 1, (n_points, 1))
return (torch.tensor(x, dtype=torch.float32, device=self.problem.device),
torch.tensor(t, dtype=torch.float32, device=self.problem.device))
def _hammersley_sequence(self, n_points, dim):
"""Generate Hammersley sequence"""
points = np.zeros((n_points, dim))
points[:, 0] = np.linspace(0, 1, n_points)
for d in range(1, dim):
base = self._nth_prime(d)
points[:, d] = self._van_der_corput(np.arange(n_points), base)
return points
def _van_der_corput(self, n, base):
"""Generate Van der Corput sequence for given base"""
seq = np.zeros_like(n, dtype=float)
denom = 1
while np.any(n > 0):
seq += (n % base) / (base * denom)
n = n // base
denom *= base
return seq
def _nth_prime(self, n):
"""Get nth prime number"""
primes = [2]
num = 3
while len(primes) <= n:
is_prime = True
for p in primes:
if p * p > num:
break
if num % p == 0:
is_prime = False
break
if is_prime:
primes.append(num)
num += 2
return primes[n]
def _qmc_points(self, n_points, method):
if method == 'hammersley':
samples = self._hammersley_sequence(n_points, 2)
else:
sampler = {
'lhs': qmc.LatinHypercube(d=2),
'halton': qmc.Halton(d=2),
'sobol': qmc.Sobol(d=2)
}[method]
samples = sampler.random(n=n_points)
x = 2 * samples[:, 0:1] - 1
t = samples[:, 1:2]
return (torch.tensor(x, dtype=torch.float32, device=self.problem.device),
torch.tensor(t, dtype=torch.float32, device=self.problem.device))
class DiffusionSolver:
"""Main solver class that handles training and visualization"""
def __init__(self, problem):
self.problem = problem
self.point_generator = PointGenerator(problem)
def train(self, n_points, sampling_method, n_epochs=15000):
"""Train PINN with given number of points and sampling method"""
model = PINN(self.problem)
optimizer = torch.optim.Adam(model.parameters(), lr=0.001)
# Generate training points
x_interior, t_interior = self.point_generator.generate_points(n_points, sampling_method)
# Generate boundary points
n_boundary = 200
x_boundary = torch.linspace(-1, 1, n_boundary, device=self.problem.device).reshape(-1, 1)
t_boundary = torch.zeros(n_boundary, 1, device=self.problem.device)
# Add corner points
corners_x = torch.tensor([-1., -1., 1., 1.], device=self.problem.device).reshape(-1, 1)
corners_t = torch.tensor([0., 1., 0., 1.], device=self.problem.device).reshape(-1, 1)
pbar = tqdm(range(n_epochs), desc=f'Training ({sampling_method}, {n_points} points)')
for epoch in pbar:
optimizer.zero_grad()
residual = model.compute_pde_residual(x_interior, t_interior)
pde_loss = torch.mean(residual**2)
u_initial = model(x_boundary, t_boundary)
ic_loss = torch.mean((u_initial - self.problem.exact_solution(x_boundary, t_boundary))**2)
t_bc = torch.linspace(0, 1, n_boundary, device=self.problem.device).reshape(-1, 1)
u_left = model(torch.ones_like(t_bc, device=self.problem.device) * -1, t_bc)
u_right = model(torch.ones_like(t_bc, device=self.problem.device), t_bc)
bc_loss = torch.mean(u_left**2 + u_right**2)
u_corners = model(corners_x, corners_t)
corner_loss = torch.mean(u_corners**2)
loss = pde_loss + 10.0 * ic_loss + 10.0 * bc_loss + 10.0 * corner_loss
loss.backward()
optimizer.step()
if epoch % 100 == 0:
pbar.set_postfix({'loss': f'{loss.item():.2e}'})
return model, self.compute_error(model)
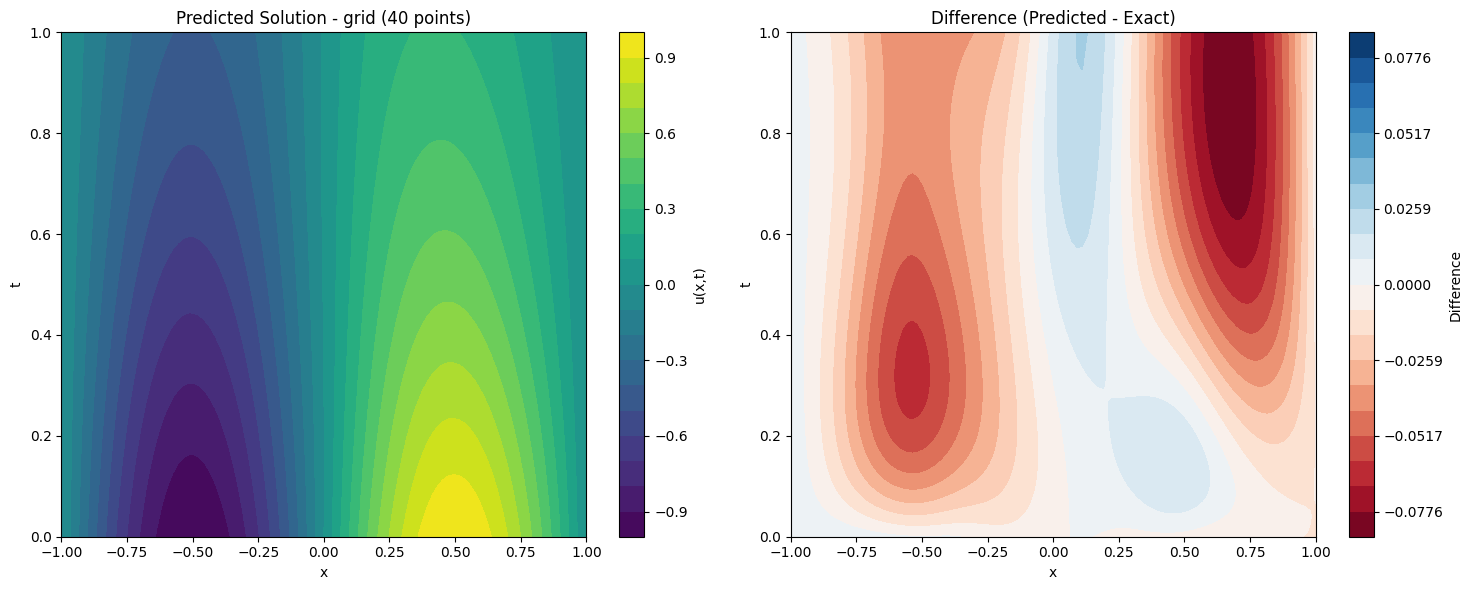
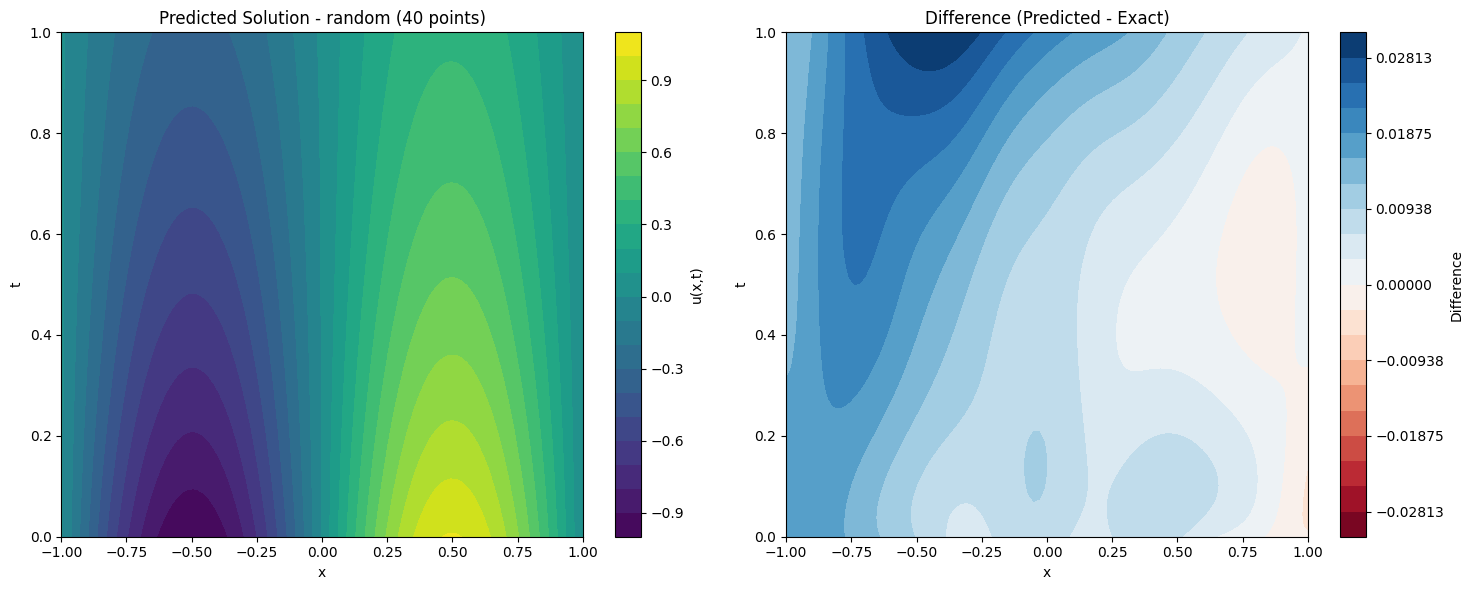
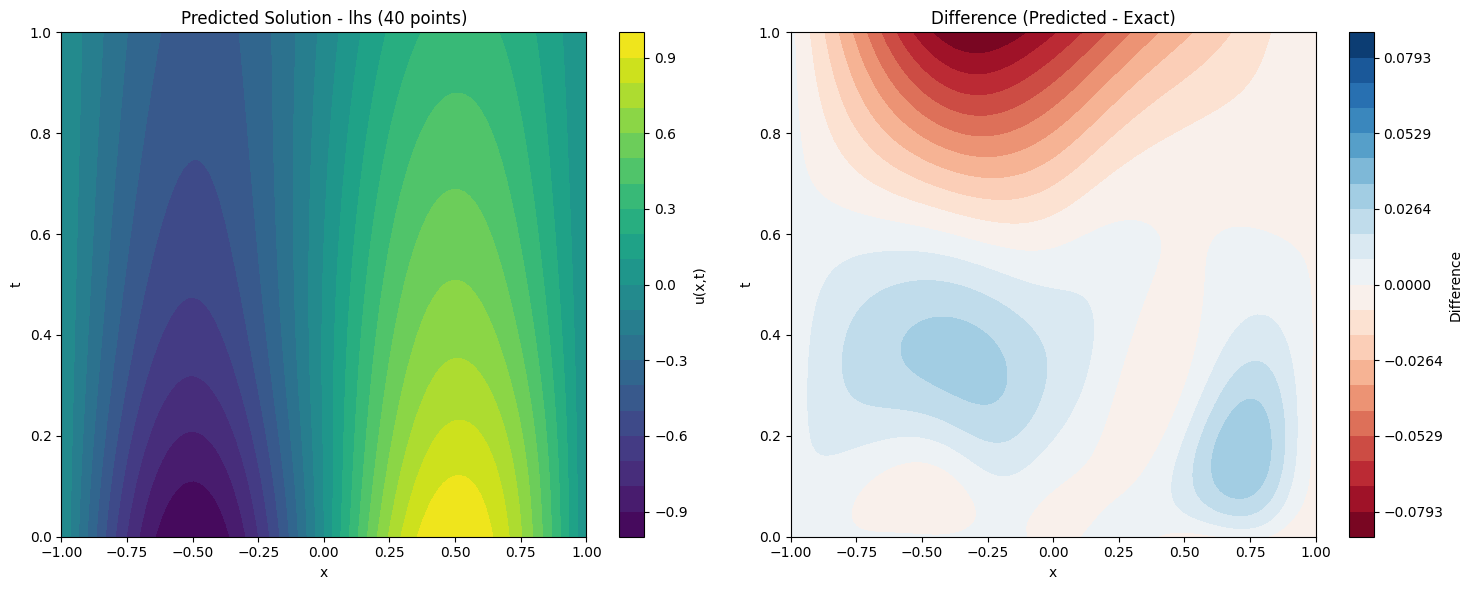
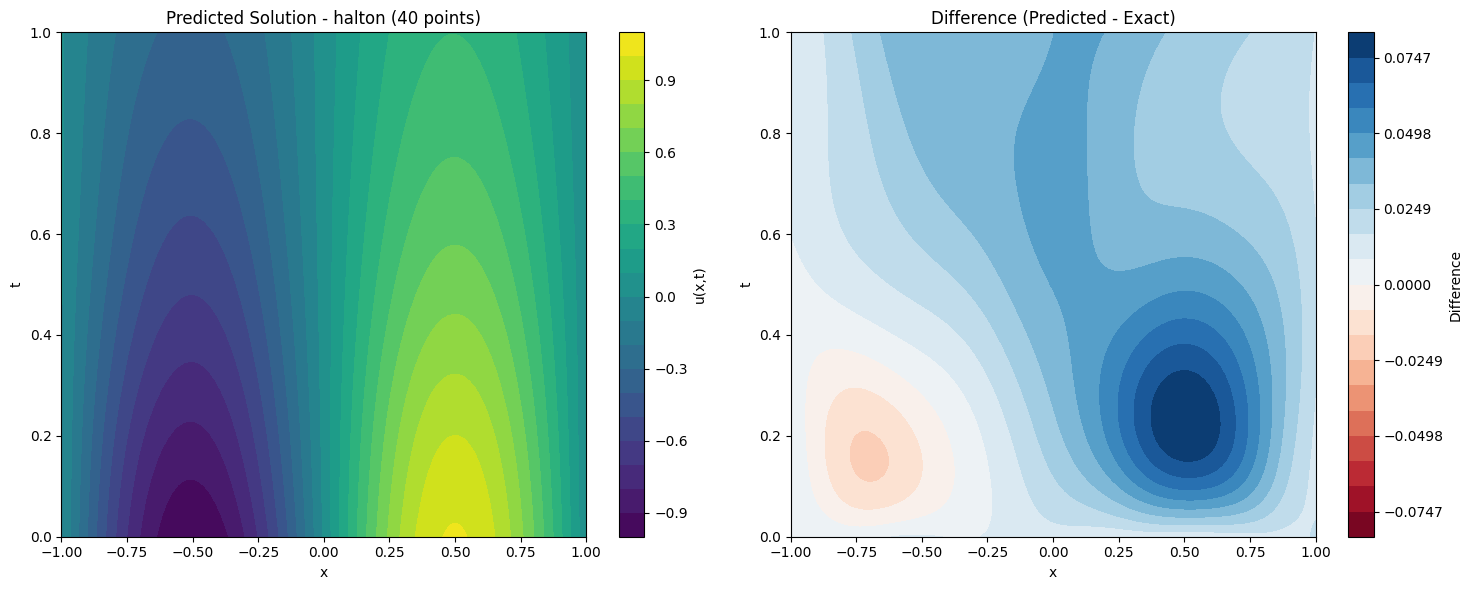
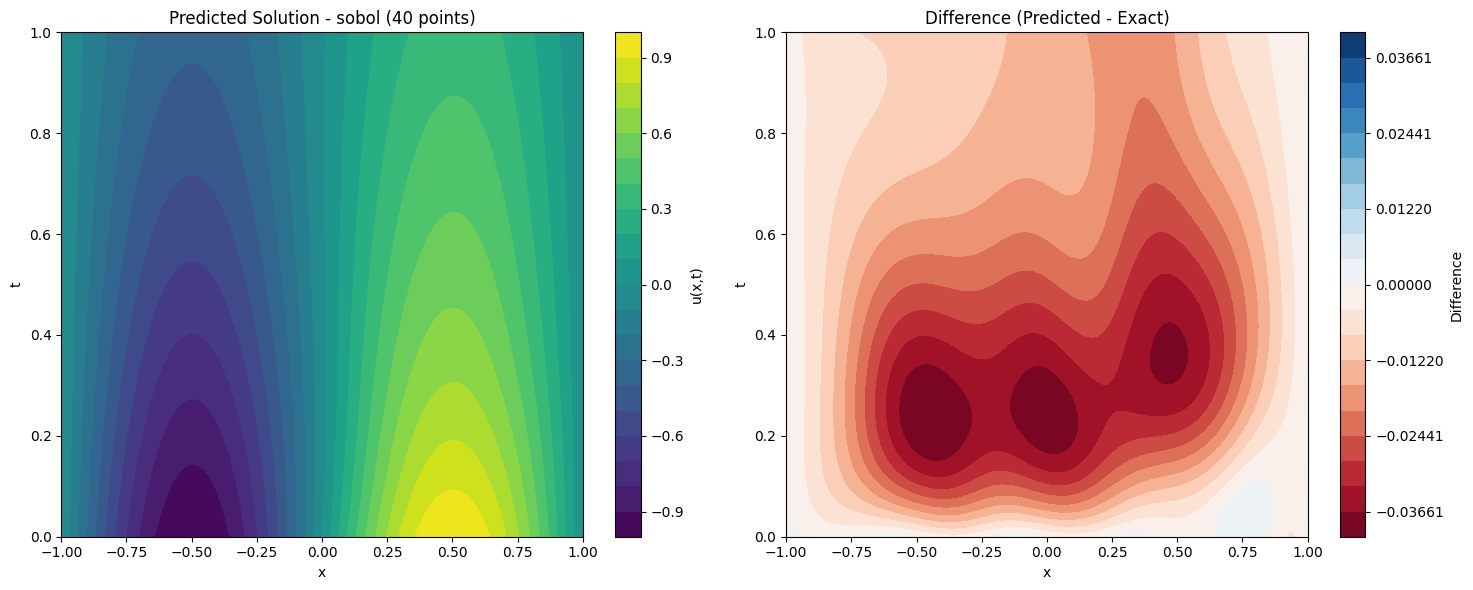
def plot_solution_comparison(self, model, method, n_points, n_test=100):
"""Plot predicted solution and its difference from exact solution"""
x = torch.linspace(-1, 1, n_test, device=self.problem.device)
t = torch.linspace(0, 1, n_test, device=self.problem.device)
X, T = torch.meshgrid(x, t, indexing='ij')
X_flat = X.reshape(-1, 1)
T_flat = T.reshape(-1, 1)
with torch.no_grad():
pred = model(X_flat, T_flat).reshape(n_test, n_test).cpu().numpy()
exact = self.problem.exact_solution(X_flat, T_flat).reshape(n_test, n_test).cpu().numpy()
diff = pred - exact
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(15, 6))
# Predicted solution
im1 = ax1.contourf(X.cpu().numpy(), T.cpu().numpy(), pred, levels=20, cmap='viridis')
ax1.set_xlabel('x')
ax1.set_ylabel('t')
ax1.set_title(f'Predicted Solution - {method} ({n_points} points)')
plt.colorbar(im1, ax=ax1, label='u(x,t)')
# Difference plot
max_diff = np.max(np.abs(diff))
levels = np.linspace(-max_diff, max_diff, 21)
im2 = ax2.contourf(X.cpu().numpy(), T.cpu().numpy(), diff, levels=levels, cmap='RdBu')
ax2.set_xlabel('x')
ax2.set_ylabel('t')
ax2.set_title('Difference (Predicted - Exact)')
plt.colorbar(im2, ax=ax2, label='Difference')
plt.tight_layout()
plt.show()
print(f"Maximum absolute difference: {np.max(np.abs(diff)):.2e}")
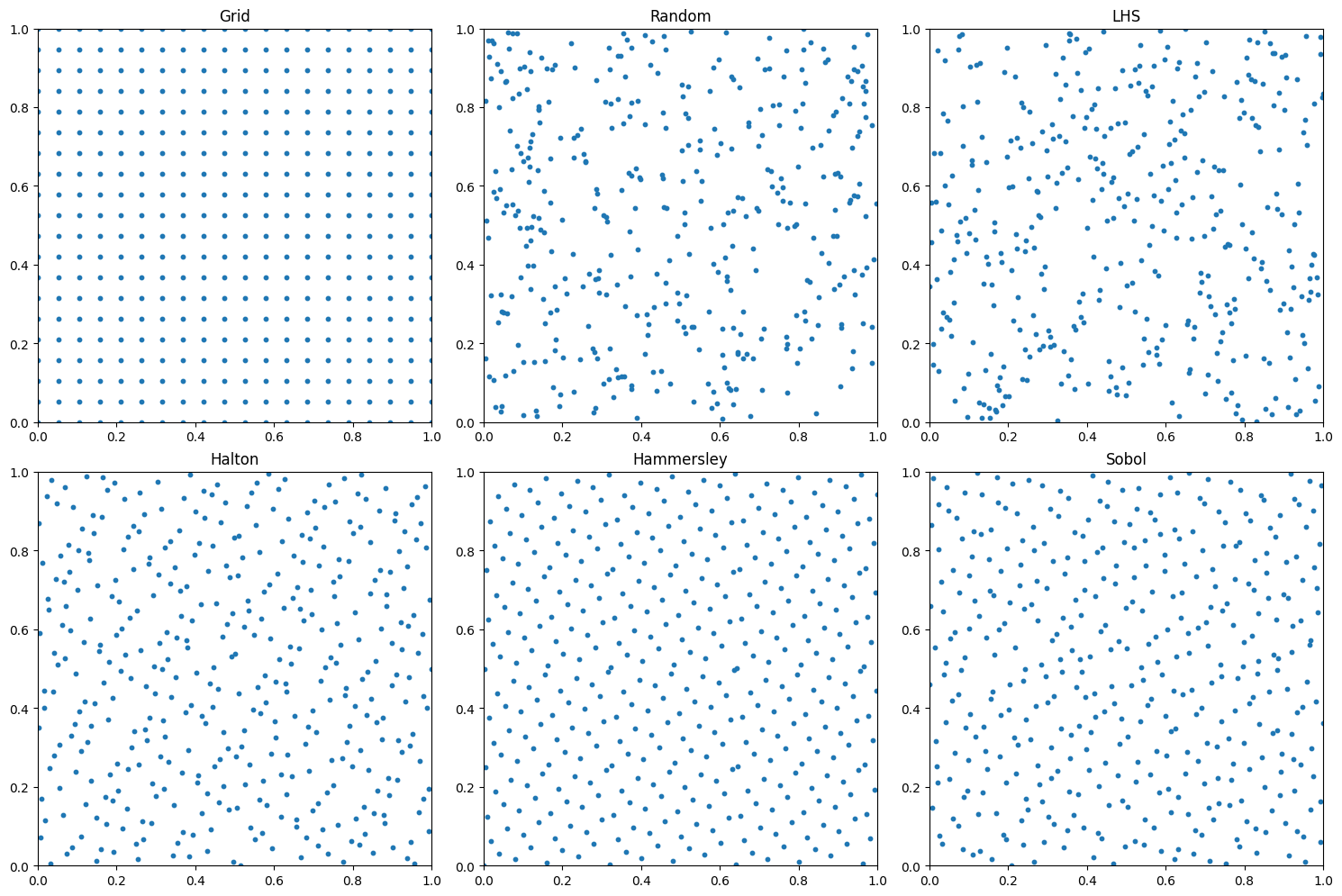
def plot_collocation_points(self, n_points, methods):
"""Plot collocation points for different sampling methods"""
n_cols = 3
n_rows = (len(methods) + n_cols - 1) // n_cols
# fig, axes = plt.subplots(n_rows, n_cols, figsize=(20, 10))
# axes = axes.flatten()
subplot_size = 6 # Size of each square subplot
fig_width = n_cols * subplot_size
fig_height = n_rows * subplot_size
fig, axes = plt.subplots(n_rows, n_cols, figsize=(fig_width, fig_height))
axes = axes.flatten()
# Common parameters
n_boundary = 20
boundary_x = np.linspace(-1, 1, n_boundary)
boundary_t = np.zeros_like(boundary_x)
t_bc = np.linspace(0, 1, n_boundary)
left_x = np.full_like(t_bc, -1)
right_x = np.full_like(t_bc, 1)
corners_x = [-1, -1, 1, 1]
corners_t = [0, 1, 0, 1]
handles, labels = None, None
for idx, method in enumerate(methods):
ax = axes[idx]
# Generate collocation points
x_interior, t_interior = self.point_generator.generate_points(n_points, method)
# Plot interior points
scatter1 = ax.scatter(x_interior.cpu(), t_interior.cpu(), c='blue', alpha=0.6, s=30, label='Interior points')
# Plot boundary points
scatter2 = ax.scatter(boundary_x, boundary_t, c='red', alpha=0.6, s=80, marker='*', label='Initial condition')
# Plot periodic boundary points
scatter3 = ax.scatter(left_x, t_bc, c='green', alpha=0.6, marker='+', s=80, label='Boundary condition')
ax.scatter(right_x, t_bc, c='green', alpha=0.6, marker='+', s=80)
# Plot corner points
scatter4 = ax.scatter(corners_x, corners_t, c='purple', s=100, alpha=0.8, marker='^', label='Corner points')
# Set labels and title
ax.set_xlabel('x')
ax.set_ylabel('t')
ax.set_title(f'{method.upper()} Sampling')
ax.grid(True, alpha=0.3)
ax.set_xlim(-1.1, 1.1)
ax.set_ylim(-0.1, 1.1)
#ax.set_aspect('equal') # Make subplot square
# Increase tick label sizes
ax.tick_params(axis='both', labelsize=12)
# Icnrease font size of title
ax.title.set_fontsize(16)
# Store handles and labels from first plot
if idx == 0:
handles = [scatter1, scatter2, scatter3, scatter4]
labels = [h.get_label() for h in handles]
# Remove empty subplots
for idx in range(len(methods), len(axes)):
fig.delaxes(axes[idx])
# Add legend below all subplots with larger font
fig.legend(handles, labels, loc='center', bbox_to_anchor=(0.5, 0),
ncol=4, borderaxespad=1, fontsize=16)
plt.tight_layout()
plt.show()
def compute_error(self, model, n_test=1000):
"""Compute L2 relative error"""
x_test = torch.linspace(-1, 1, n_test, device=self.problem.device).reshape(-1, 1)
t_test = torch.linspace(0, 1, n_test, device=self.problem.device).reshape(-1, 1)
X_test, T_test = torch.meshgrid(x_test.squeeze(), t_test.squeeze(), indexing='ij')
x_test = X_test.reshape(-1, 1)
t_test = T_test.reshape(-1, 1)
with torch.no_grad():
u_pred = model(x_test, t_test)
u_exact = self.problem.exact_solution(x_test, t_test)
return (torch.norm(u_pred - u_exact) / torch.norm(u_exact)).item()
def main():
# Initialize problem and solver
problem = DiffusionProblem()
solver = DiffusionSolver(problem)
# Define sampling methods and points to test
sampling_methods = ['grid', 'random', 'lhs', 'halton', 'sobol', 'hammersley']
n_points_range = [10, 20, 40, 60, 80]
# Plot collocation points comparison
solver.plot_collocation_points(80, sampling_methods)
# Store results
results = {method: [] for method in sampling_methods}
models = {}
# Training loop
for n_points in n_points_range:
for method in sampling_methods:
model, error = solver.train(n_points, method)
results[method].append(error)
# Store model for visualization (middle point count)
if n_points == 20:
models[method] = model
# Plot error comparison
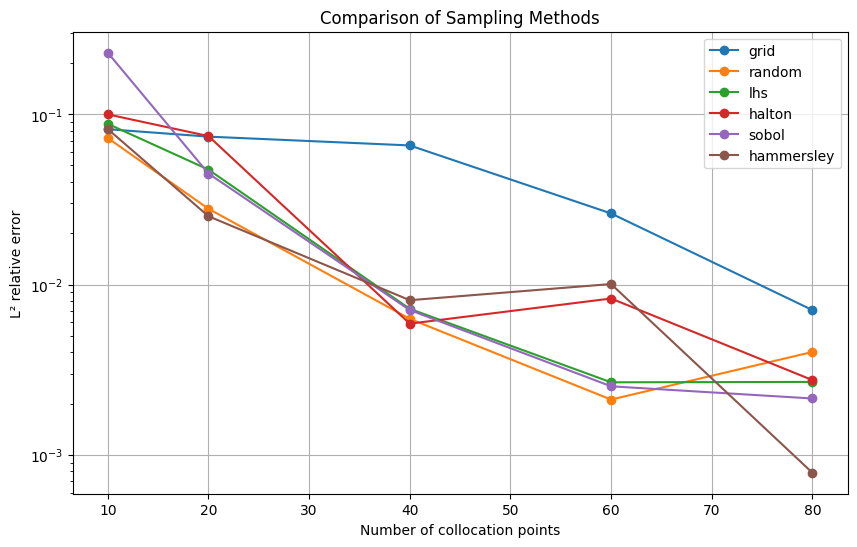
plt.figure(figsize=(10, 6))
for method in sampling_methods:
plt.plot(n_points_range, results[method], label=method, marker='o')
plt.yscale('log')
plt.xlabel('Number of collocation points')
plt.ylabel('L² relative error')
plt.legend()
plt.grid(True)
plt.title('Comparison of Sampling Methods')
plt.show()
# Plot solution comparisons for each method
for method, model in models.items():
solver.plot_solution_comparison(model, method, 40)
# Print final errors
print("\nFinal L² errors:")
for method in sampling_methods:
print(f"{method:10s}: {results[method][-1]:.2e}")
if __name__ == '__main__':
main()